Contributors: Ela Dudek, Paige Grant, Laine Lysyk, Tautvydas Paskevicius, Simon Wu
The development of recombinant proteins stands out as a major milestone in biotechnology over the past century. These proteins are synthesized within different types of platforms, known as expression or heterologous hosts, which can be either eukaryotic or prokaryotic. Commonplace platforms today are bacteria, yeast, insect, and mammalian cells. With advancements in omics data and genetic engineering tools, scientists can now modify these hosts artificially to enhance the production of recombinant proteins on a large scale. This has led to the widespread use of recombinant proteins across various industries, with the global market expected to exceed USD 2.4 billion by 2027.
Protein expression is a complex task; the whole process from transcription to translation involves hundreds of components and many variables that are cross‐correlated. The more difficult a protein is to express, the more crucial it becomes to assess the strengths and weaknesses of the heterologous hosts. Current expression systems are limited in what they can produce, necessitating the search for alternative platforms.
Before selecting the optimal host, we must understand the different challenges that difficult proteins present.
Protein targets are becoming increasingly difficult to express; more than 50% of recombinant protein production processes fail at the expression stage. These challenging proteins fall into the classification of difficult-to-express proteins (DTEPs).
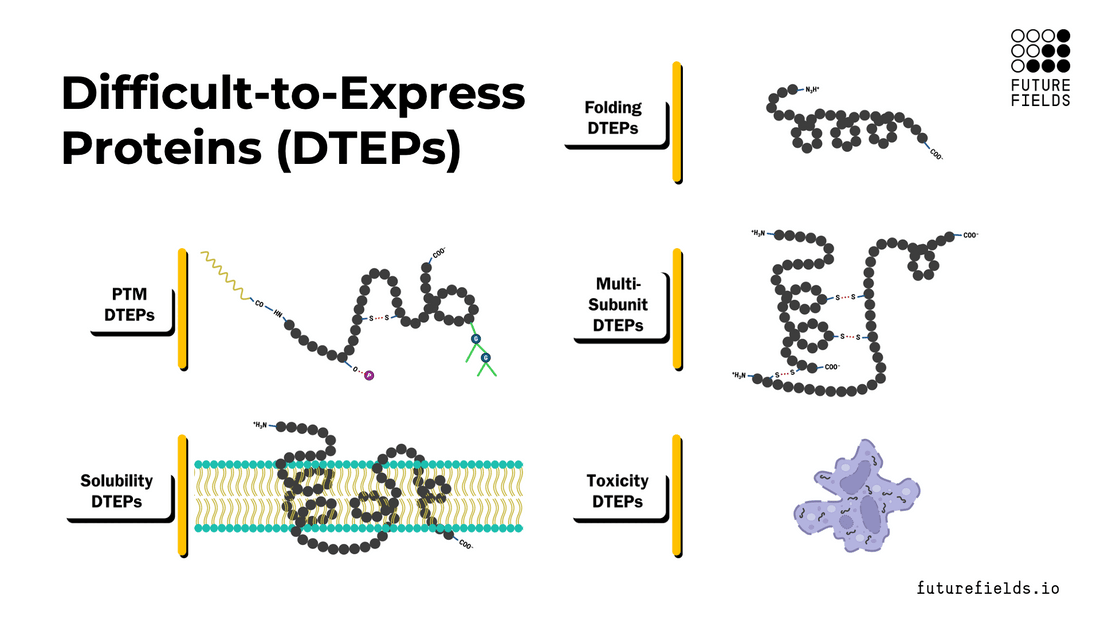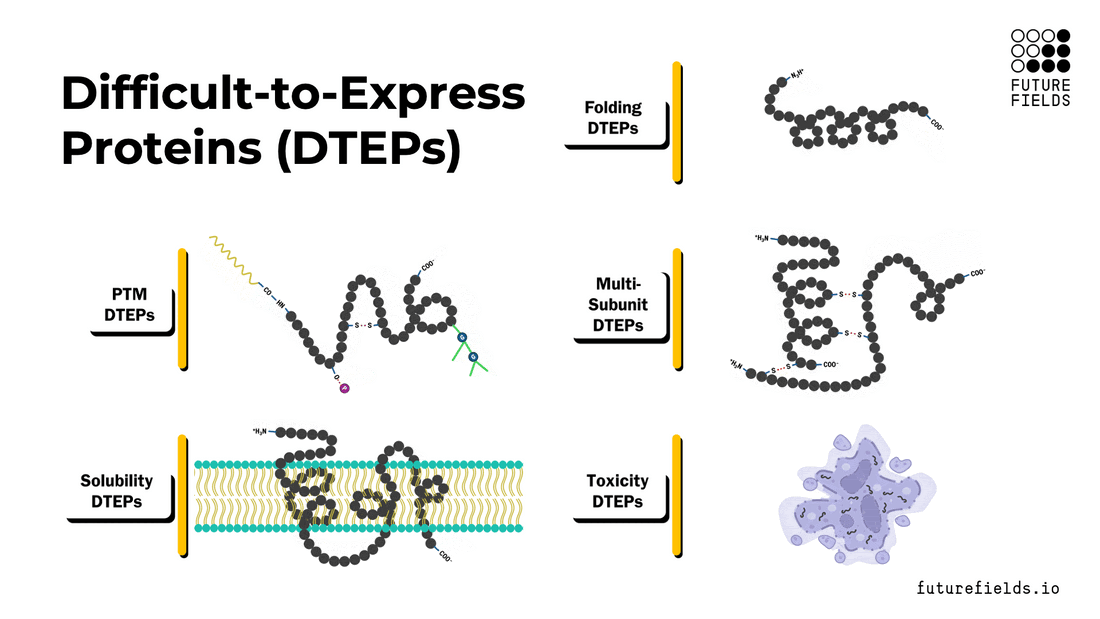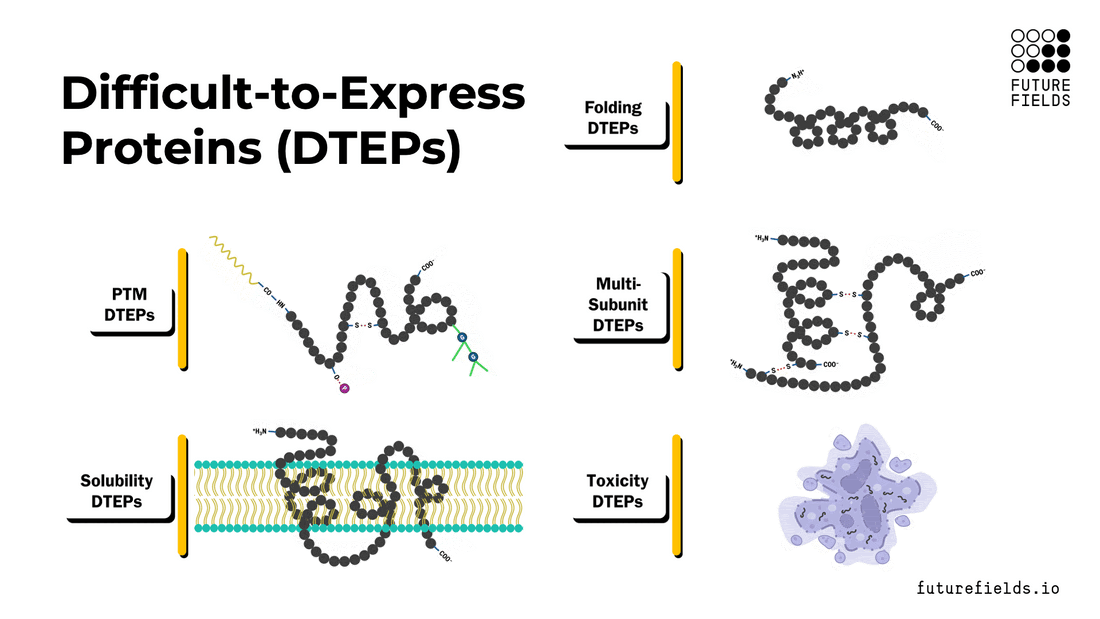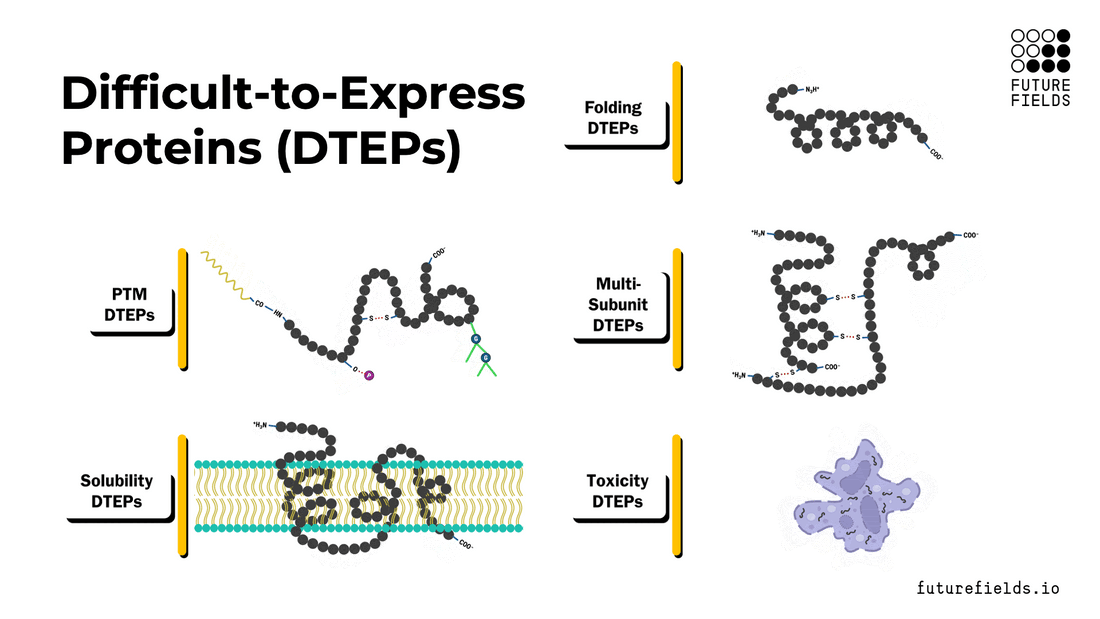
What are difficult-to-express proteins (DTEPs)?
Broadly, DTEPs can be characterized with a spectrum of intrinsic properties that make a specific protein difficult to express and/or purify. These properties include folding, post-translational modifications, multi-subunit complex assembly, solubility, and toxicity. A target protein can possess all these properties to a varying degree, creating a range of difficulties for successful recombinant protein production.
In this article, we will explore each property, its relationships with other properties, and the challenges it poses to the scientist and the protein expression system.
Protein Folding (and Misfolding)
Every protein is initially expressed as an unstructured, linear chain of amino acids, usually described as random-coil. In order for a protein to acquire its native and functional three-dimensional shape, each polypeptide must be properly folded.
In the cell, this folding process is assisted by molecular chaperones that transiently bind and stabilize folding intermediates, often through exposed hydrophobic patches. These chaperones prevent protein misfolding and aggregation, which can be toxic to the cell. Chaperones also ensure the balance in cellular proteostasis. Certain proteins with complex topological structures are intrinsically difficult to fold and require prolonged contact time with chaperones, resulting in significantly lower folding rates.
In addition to molecular chaperones, many other factors influence the folding capacity and efficiency, such as the solvent or milieu polarity, concentration of salts, presence of cofactors, redox potential, pH, and temperature. Protein folding is fundamental in that it can affect or be affected by other properties of DTEPs. For example, prolonged incorrect folding, can lead to a buildup of protein misfolding that damages cells, causing several abnormalities. Failure of a protein to fold into the correct 3D structure typically results in the production of inactive proteins, but it may also trigger modified and/or toxic functionalities.
On the other hand, protein folding is heavily impacted by post translational modifications, or PTMs. As “regulators of protein folding”, PTMs can contribute to both the proper folding and misfolding of proteins. A specific positive example is a disulfide bond formation, a critical PTM that contributes to the folding, structural integrity, stability, and function of many proteins.
Protein Post-Translational Modifications (PTMs)
Protein post-translational modifications (PTMs) represent a vast array of covalent alterations that significantly contribute to protein structural and functional diversity.
With over 400 different types of PTMs identified to date, these modifications play pivotal roles in various aspects of protein biogenesis and function. Among the extensively studied PTMs are glycosylation, phosphorylation, ubiquitination, acylation, methylation, nitration, and acetylation. While each PTM type serves a distinct function, glycosylation stands out as the most prevalent modification observed in therapeutic proteins.
Following protein synthesis within a cell, PTMs often occur, profoundly influencing protein structure, function, and localization. These modifications involve the addition of chemical groups–such as phosphate, acetyl, or methyl groups–to specific amino acids, or the cleavage of certain peptide bonds. For instance, phosphorylation serves as a molecular switch, regulating protein activity in response to cellular signals. Glycosylation plays essential roles in protein folding, stability, and cell-cell recognition, while acetylation and methylation can impact gene expression by modifying histone proteins associated with DNA. Additionally, ubiquitination marks proteins for degradation by the proteasome, controlling their cellular levels.
Understanding the complexity of PTMs is crucial for optimizing the expression of recombinant proteins in various heterologous hosts. While multiple prokaryotic and eukaryotic-based expression systems have been developed, the careful choice of expression system for producing glycosylated recombinant proteins is crucial since it can significantly impact their glycosylation patterns, which in turn can affect their biological activity, stability and immunogenicity. Also, the intrinsic complexity of PTMs presents additional challenges during recombinant protein expression. PTM heterogeneity can make it challenging to separate target protein molecules with different or partial modifications, impacting protein purity and yield. Moreover, PTMs can influence protein solubility, affecting protein stability and half-life.
Addressing these challenges requires a comprehensive understanding of PTMs and their impact on recombinant protein expression. By leveraging innovative strategies and advancements in expression systems, researchers can overcome hurdles associated with PTMs and unlock the full potential of biologically and therapeutically significant molecules. Continued exploration of PTMs holds promise for yielding insights into cellular biology and facilitating the development of novel therapeutic interventions.
Multi-Subunit Complexes
Protein-protein interactions are key for the functionality of most proteins. Some of these interactions are transient, between two separate proteins with individual functions, while others are structural components of a single functional protein. In these cases, the individual amino acid chains are referred to as subunits, and the protein itself is referred to as a multi-subunit protein complex or oligomer. Multi-subunit proteins present unique challenges for protein expression, as the subunits finding each other and then assembling together correctly are both additional complex steps in the process of correctly forming a protein.
Subunit assembly can occur simultaneously with protein folding, or after the individual subunits are formed. Often, when folding occurs simultaneously, these interactions are obligatory, meaning the individual subunits are not stable alone. Conversely, when folding precedes the assembly of the subunits, the interactions are non-obligatory, meaning the subunits are stable as monomers. Either of these situations can pose problems for expression. In either case, expression must be sufficiently high for subunits to quickly and easily locate one another. Otherwise, for obligatory subunits, misfolding and aggregation will occur, while for non-obligatory subunits, stable but inactive monomers will be the dominant species.
Many multi-subunit complexes are homomeric, meaning they consist of multiple copies of the same subunit. One estimate suggests that 30–50% of all proteins assemble into homomers. These require only the expression of one amino acid chain, simplifying expression challenges somewhat compared to heteromers. However, monomers or oligomers with an incorrect number of subunits are difficult to separate from the correctly formed protein, as many of the intrinsic properties of the correctly formed protein may be shared with the incorrectly assembled forms (pI, hydrophobicity, affinity). Heteromeric complexes, which contain subunits that are different from each other, offer a further challenge still, as more than one subunit needs to express simultaneously. The correct proportion of each subunit needs to be produced for correct assembly, which is a technically difficult task in genetic engineering. As oligomers become larger, with more subunits, this becomes more difficult, and the burden on the host increases.
The most commonly produced biologics in the biopharmaceutical industry are monoclonal antibodies, which are multi-subunit proteins. This global market is projected to continue to grow at a CAGR of 12.2%, surpassing small molecules in the next five years. As such, effective methods for expressing oligomeric proteins are important for both research and for pharmaceuticals.
Insoluble Proteins
Protein solubility is a critical aspect of protein production and purification processes, influencing both the yield and quality of proteins. Solubility refers to the ability of a protein to dissolve in a solvent, typically water, to form a homogeneous solution. The solubility of a protein is governed by its amino acid composition, structure, and environmental conditions such as pH, temperature, and ionic strength. Hydrophilic residues promote solubility, while hydrophobic residues tend to induce protein aggregation and precipitation. Many proteins exhibit poor solubility when expressed heterologously in recombinant systems; improper processing, misfolding, and hydrophobic mismatch can all result in poor solubility due to hydrophobic patches of a protein being exposed to solution. Insoluble proteins can form inclusion bodies, aggregated protein structures that are often difficult to solubilize and refold into their native conformation, often rendering the protein non-functional. Inclusion body formation not only reduces protein yield but also complicates downstream purification and functional studies.
The class of proteins that present the most challenges in regards to solubility are transmembrane proteins. Transmembrane proteins are intrinsically hydrophobic as they reside embedded within cellular membranes. During expression of transmembrane proteins, large surface hydrophobic regions that would natively be protected within a lipid bilayer may become exposed if not stabilized by chaperones during folding or before translocation and insertion into a suitable membrane environment. These hydrophobic regions hinder solubility and increase the propensity for misfolding and aggregation.
In recombinant hosts, the cellular machinery that translocates transmembrane proteins into cellular membranes can easily become overwhelmed during expression, leading these hydrophobic proteins to either aggregate in the cytosol or incorrectly incorporate into the membrane. In addition to the challenges with membrane insertion, many transmembrane proteins have complex folding that requires multi-subunit association within the lipid bilayer. Furthermore, membrane proteins often require specific lipid interactions to mediate function which may not be present in the recombinant host.
The production of recombinant membrane proteins is further complicated by the need to isolate the protein into a suitable environment that protects the hydrophobic regions of the protein from exposure to solution, whether through the use of detergents or membrane-mimetic systems such as proteoliposomes, while also maintaining stability of the protein in its native conformation so that it retains functionality.
Toxic Proteins
The principle that "the dose makes the poison" holds true when considering the toxicity of proteins. While different proteins exhibit varying degrees of toxicity, any recombinant protein, when expressed at high levels, can pose a threat to the host organism. This toxicity often arises from the hijacking of cellular machinery or the overburdening of the host system, impairing its normal physiological functions. However, proteins that are particularly challenging to express due to their toxicity typically exert their harmful effects through their intrinsic activities.
Enzymes, which are active proteins that catalyze biochemical reactions, may be toxic to the host for this reason. The enzymatic activity may be incompatible with the host or unsuitable for the host's environment. For example, aberrant activity of a kinase, which phosphorylates proteins, may interfere with the intrinsic signaling pathways of the host. Mitigating protein toxicity is crucial for enhancing production yields, whether through minimizing exposure to high protein concentrations or engineering cell lines to withstand the effects of toxic proteins, thereby promoting growth and optimizing production efficiency.
Overlapping Properties of DTEPs
Difficult-to-express proteins rarely possess a single challenging property. Proteins can simultaneously be difficult to fold, have PTMs and/or fold into multi-subunit complexes, and display toxicity and/or solubility issues. These challenges can also exist in varying levels of severity.
Instead of tackling each property in isolation, one useful way to approach a DTEP target is to map it out across properties in relation to other DTEP targets.
For example, a transmembrane protein’s hydrophobicity not only hinders solubility, but also increases the likelihood of misfolding and aggregation. Certain transmembrane proteins may also require specific PTMs and multi-subunit association for function. Monoclonal antibodies are multi-subunit proteins with inherently complex folding, and having the correct PTMs is important for their function. Toxic proteins can be intrinsically toxic, despite seeming simple based on their PTMs, folding, solubility, and lack of multi-subunit complex formation. The opposite may also be true–high toxicity could result from misfolding, insolubility, PTMs, and/or incorrectly formed multi-subunit complexes.
Understanding that DTEPs have multiple challenging properties, selecting the optimal expression host then becomes an evaluation of whether or not it can effectively address multiple challenges. A one-size-fits-all approach that relies on conventional expression systems will not suffice. These proteins require an alternative, robust, and customizable platform.
At Future Fields, we understand that unconventional proteins require unconventional solutions. This is why we have purposefully selected and refined the EntoEngine™ expression system. Using Drosophila melanogaster, we can express even the most challenging DTEPs by leveraging the built-in, robust expression capabilities of eukaryotes; a century of innovative genetic research; and a diverse set of tissue types that allows for parallel protein expression strategies to optimize yield.
Future Fields’ EntoEngine™ is a powerful and versatile expression platform that can play a crucial role in unlocking the full research and therapeutic potential of complex protein expression challenges.
Expressing Your Difficult-to-Express Protein
Difficult-to-express proteins encompass a diverse range of molecules, including membrane proteins, multidomain proteins, and proteins with intricate post-translational modification requirements. These proteins are notorious for a multitude of challenges, including their low expression levels, misfolding tendencies, poor solubility, and potential cell toxicity issues. This makes their production in heterologous systems like E. coli and yeast particularly challenging. Therefore, current conventional recombinant protein expression systems often struggle to adequately produce DTEPs.
Overcoming these hurdles typically requires tailored, protein-specific solutions that often involve laborious efforts for gene and protein engineering, co-expression of chaperones, and optimization of cultivation conditions. Future Fields’ EntoEngine™ is a powerful and versatile expression platform that offers these solutions, unlocking the full research and therapeutic potential of scientists creating complex, life-changing proteins.
Contact us today to learn more about expressing your difficult protein.
Future Fields produces difficult-to-express proteins with its novel Dropsophila-based platform: the EntoEngine™.
The EntoEngine™ is the world’s first synthetic biology system to use fruit flies for recombinant protein production.
Sources
- Khoury, G. A., Baliban, R. C., & Floudas, C. A. (2011). Proteome-wide post-translational modification statistics: Frequency analysis and curation of the Swiss-Prot database. Scientific Reports, 1, 90. DOI: 10.1038/srep00090
- Ramazi, S., & Zahiri, J. (2021). Post-translational modifications in proteins: Resources, tools and prediction methods. Database, 2021. DOI: 10.1093/database/baab080
- Lim, B. K., Lim, C. S., Remus, D. M., Chen, A., van Dolleweerd, C., & Gardner, P. P. (2021). Analysis of 11,430 recombinant protein production experiments reveals that protein yield is tunable by synonymous codon changes of translation initiation sites. PLoS Computational Biology, 17(10). DOI: 10.1371/journal.pcbi.1009400
- Yang, A., Cho, K., & Park, H.-S. (2018). Chemical biology approaches for studying posttranslational modifications. RNA Biology, 15(4-5), 427–440. DOI: 10.1080/15476286.2017.1360468
- Walsh, G. (2010). Post-translational modifications of protein biopharmaceuticals. Drug Discovery Today, 15(17-18), 773-780.
- Qin, Y., et al. (2016). EcoExpress: Highly efficient construction and expression of multicomponent protein complexes in Escherichia coli. ACS Synthetic Biology, 5(11), 1239-1246.
- Levy, E. D., & Teichmann, S. A. (2013). Structural, evolutionary, and assembly principles of protein oligomerization. Progress in Molecular Biology and Translational Science, 117, 25-51.
- Hegde, R. S., & Keenan, R. J. (2022). The mechanisms of integral membrane protein biogenesis. Nature Reviews Molecular Cell Biology, 23, 107–124. DOI: 10.1038/s41580-021-00413-2
- Cancer Monoclonal Antibodies Market Size Estimated to Reach USD 410.95 Billion by 2032. (n.d.). Retrieved from https://www.biospace.com/article/releases/cancer-monoclonal-antibodies-market-size-estimated-to-reach-usd-410-95-billion-by-2032/
- Small Molecules, Big Market Opportunities. (n.d.). Retrieved from https://manufacturingchemist.com/small-molecules-big-market-opportunities-210051